| |
 |
 |
| |
| Home | Project | People | Research | Education | Archive | |
 |
Environmental Genomics |
[English | Japanese ] |
 |

 |
![Chemical Genomics]](../img/btn_e_chemo_01.gif)  |
 |
Minoru Kanehisa
Professor,
Bioinformatics Center,
Institute for Chemical Research, Kyoto University |
Integration of Genomics and Chemistry by Reaction Networks
The macromolecular structures of DNAs, RNAs, and proteins are determined by template-based syntheses of replication, transcription, and translation, which represent the processes of genetic information transmission and expression as shown by the central dogma of molecular biology. In contrast, the structures of carbohydrates, lipids, and various secondary metabolites are determined, not by template structures, but by biosynthetic pathways. Such biosynthetic codes are far more complex than genetic codes, and our knowledge is still quite limited. Bacteria and plants are known to produce diverse substances, many of which have medical and pharmaceutical relevance, including antibiotics and crude drugs. With the complete genome sequences becoming available for an increasing number of organisms, it should in principle become possible to infer a complete set of biochemical substances produced by each organism, and furthermore to infer a set of xenobiotic substances that are degraded by an organism. We are thus developing new bioinformatics approaches for integrated analyses of genomic and chemical (environmental) information by organizing our knowledge on biosynthetic and biodegradation pathways, which will be used in practical applications of drug discovery and drug design.
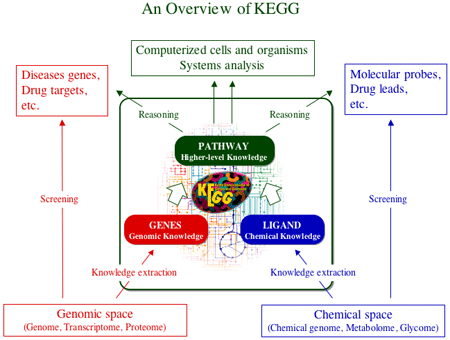
Our current knowledge on the universe of enzymatic reactions is represented by the set of EC (Enzyme Commission) numbers. The EC numbers are given to distinguish enzymatic reactions (chemical information), but they are also utilized as identifiers of enzymes and enzyme genes (genomic information). This duality of the EC numbers makes it possible to link the genomic repertoire of enzyme genes to the chemical repertoire of endogenous and exogenous substances. We have thus far developed methods to compare chemical compound structures and to classify chemical transformation patterns in enzymatic reactions, which have been used to automatically assign EC numbers given chemical structures of substrates and products. These methods enable us to convert a set of chemical compound structures to a set of enzyme genes through a network of reactions, and vice versa. In other words, the chemcal information can be used to decipher the genome, and the genomic information can be used to predict chemical structures. The results of this research project is incorporated in KEGG, a database system developed by our group and used worldwide as a knowledge information infrastructure for genome science.
 |
Top
| 2007 | 2006 | 2005 | |
|
|

