With the recent advance of experimental techniques in molecular biology, the research of modern life science is shifting to the comprehensive understanding of biological mechanisms carried out by a variety of molecules. The focus of our laboratory is placed on biological phenomena that can be represented by biological networks such as metabolic pathways and signal transduction pathways. The research objective is to develop computational techniques based on computer science and/or statistics to systematically understand the principles of biological networks at the cellular and organism level. Considering the focus of the 21st Century COE program "Knowledge Information Infrastructure for Genome Science", we here show two typical examples of our research.
New Chemical Descriptor Based-on Graph Theory
The tree-width is, in graph theory, a metric for the complexity of a graph. More precisely, it is a measure indicating how a given graph is similar to a tree. For example, if a given graph of
n nodes is a tree, its tree-width is one, and it will be two if the graph has a cycle. And it will be
n-1 if the graph is a complete graph. We checked how we can use the tree-width as a chemical descriptor for a planar chemical structure of a given chemical compound. We focused on the chemical reactions by which the tree-width of a chemical compound can change. Our first finding was that these reactions tend to have particular EC numbers. We then found that the graph connectivity of an arbitrary pathway graph changes drastically when we remove these chemical reactions. These results imply that the tree-width can be a new chemical descriptor to elucidate the structural and biochemical properties of chemical compounds.
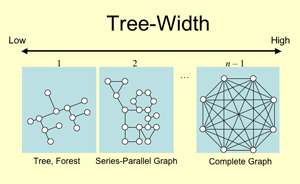 Fig. 1. Tree-Width
Fig. 1. Tree-Width
A database of metabolic pathways is a collection of chemical reactions that appeared in the literature. This indicates that each reaction is not observed under a same cellular condition, implying that a long distance path might not be true under some condition. Motivated by this doubt, we build a method to find biologically active paths in a given metabolic pathways by using microarrays. We generate a hierarchical mixture of Markov chains using a given pathway and estimate its probabilistic parameters from given microarray experiments based on an EM (Expectation-Maximization) algorithm. Experiments using real datasets showed that our method found paths actually used in terms of gene expressions as well as the correlation of chemical reactions located at distant parts in a pathway.

