 アブストラクト
アブストラクト| Date | November 18, 2011 |
| Speaker | Prof. Alexandre Varnek, Laboratory of Chemoinformatics, University of Strasbourg, France |
| Title | Chemical space: design, visualization and navigation |
| Abstract |
This presentation considers several aspects of the design of descriptor-based chemical spaces, their visualization and application to modeling and virtual screening.
1. Nonlinear Dimensionality Reduction Techniques. Various dimensionality reduction approaches will be discussed focusing mostly on Self-Organized maps (SOM) and Generative Topographic Maps (GTM) 1. The latter could be considered as a universal tool to visualize the chemical space, to predict activity profiles, to conduct virtual screening and to compare databases of chemical compounds. Unlike other popular methods of data visualization (PCA, SOM, etc), for a given molecule GTM calculates its probability to be located in the given point of rectangular 2D map. Thus, for the whole dataset GTM not only visualizes the data points, but calculates the probability density function which could be used to build structure-property models or to assess an overlap between two datasets. The model calculations performed on the DUD, GB13 and ZINK databases using the ISIDA 2 descriptors illustrate the utility of GTM. 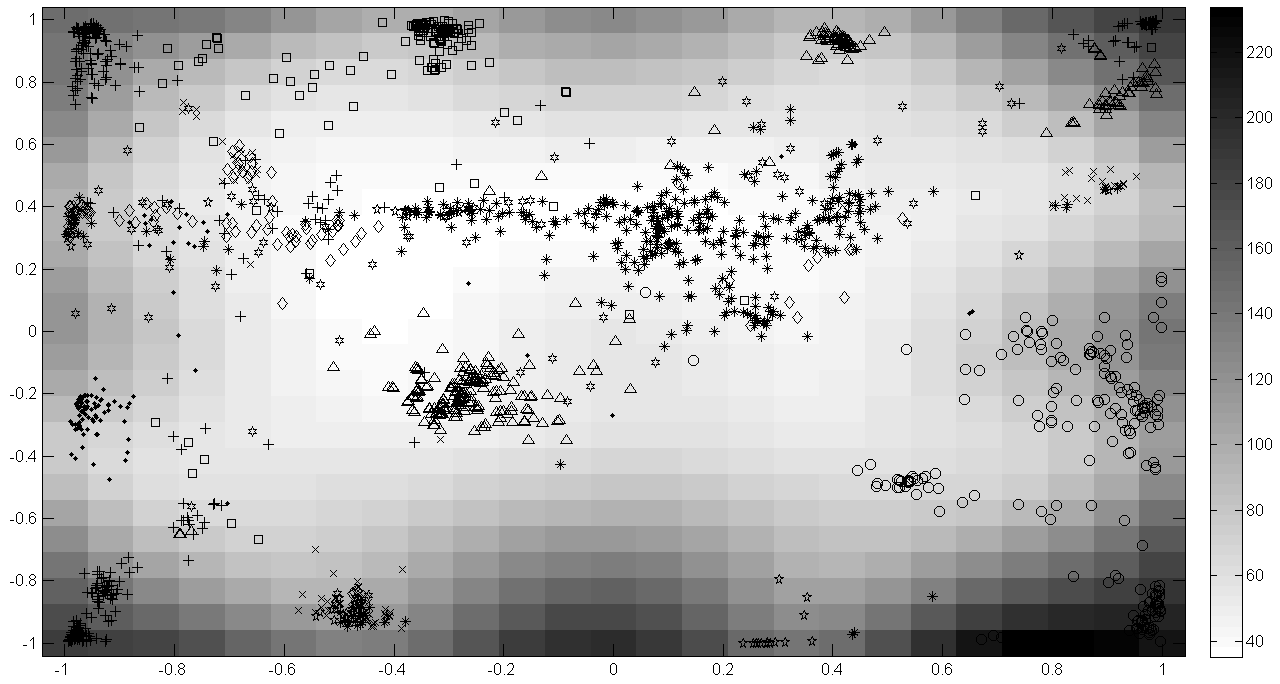 Generative Topography Map for the dataset of the DUD ligands against 10 different biological targets. The background color code corresponds to "magnification factor" which relates the distances between the objects in the initial descriptors space with those on the map. 2. Selection of optimal descriptors to design a chemical space. The Neighborhood Behavior (NB) 3 approach is an efficient method to select for a given dataset an optimal descriptors/metric combination to be used in similarity search. Here, it is illustrated on the database containing 8500 chemical reactions encoded by the Condensed Graphs of Reactions 4 from which different pools of ISIDA descriptors have been generated. The SOM and GTM based on the "best" descriptors selected in NB calculations well separate different reaction classes. 3. Acceleration of similarity search using SOM. Self-Organized Maps of the given database (DB) can be efficiently used to accelerate a similarity search if the latter is limited to the neuron to which a query molecule is projected and to selected number of the neighboring neurons. The tests performed on DB of ~ 55; 000 molecules using a set of 2000 query molecules demonstrate significant acceleration of the speed of calculations keeping reasonable screening performance. References. 1. Bishop, C.M. and M. Svensen, Neural Computation, 1998. 10(1), 215-234. 2. Varnek, A., et al., Curr. Comp.-Aid. Drug Des., 2008, 4(3), 191-198. 3. C. Koch, G. Schneider, G. Marcou, A. Varnek, D. Horvath J. Comp.-Aided Mol. Design, 2011, 25, 237-252 4. F. Hoonakker, N. Lachiche, A. Varnek, A. Wagner Int J. Artificial Intelligence Tools, 2011, 20, 253-270 |